Simulation Example
November 12, 2014
The motivation for this example has to do with different ways one can calculate individual differences with regard to the effect of some manipulation that pushes around cortisol levels.
As a general resource for simulation in R, I highly recommend Hadley Wickham’s simulation presntation. Also, the psych package has some great tools for generating data (with great support in this helpful guide by William Revelle).
library(plyr)
library(ggplot2)
library(reshape2)
Set parameters here
cortPopMeanT1 <- 10 # Mean of population
cortPopSDT1 <- 2# SD of population
cortIndivDiffMean <- 4 # Mean of true absolute difference
cortIndivDiffSD <- 1 # SD of true absolute difference
epsilonM <- 0 # Mean of error
epsilonSD <- 1 # SD of error
N <- 100 # sample size
ITERS <- 1000
expRawDiff <- cortIndivDiffMean
expPercDiff <- cortIndivDiffMean/cortPopMeanT1
1. Make a function to let us draw from some distributions without worrying about their parameterization.
We can also use this to get each person’s difference score.
someSample <- function(n,popmean,popsd,dist='gamma'){
if(dist == 'gamma'){
shape=popmean^2/popsd^2
scale=popsd^2/popmean
return(rgamma(n,shape=shape,scale=scale))
}
if(dist == 'norm'){
return(rnorm(n,mean=popmean,sd=popsd))
}
}
2. Now, let’s write a function to build our data frame
prePostDFgen <- function(n,meanInit,sdInit,meanDiff,sdDiff,eM,eSD,distInit='gamma',distDiff='norm'){
#Returns a DF with c('pre','post') %in% names(DF)
initSamp <- someSample(n,meanInit,sdInit,distInit)
diffSamp <- someSample(n,meanDiff,sdDiff,distDiff)
errSampI <- rnorm(n,eM,eSD)
errSampD <- rnorm(n,eM,eSD)
data.frame(
pre=initSamp+errSampI,
post=initSamp+diffSamp+errSampD)
}
3. Now, three functions to calculate the within-person changes
testDF<-prePostDFgen(
N,
cortPopMeanT1,
cortPopSDT1,
cortIndivDiffMean,
cortIndivDiffSD,
epsilonM,
epsilonSD,
distInit='gamma',
distDiff='norm')
rawDiff <- function(pre,post) post-pre # pre and post should be vectors
percDiff <- function(pre,post) (post-pre)/pre
residDiff <- function(pre,post) resid(lm(post~1+pre))
rawDiffMean <- function(pre,post) mean(rawDiff(pre,post))
percDiffMean <- function(pre,post) mean(percDiff(pre,post))
residDiffMean <- function(pre,post) mean(residDiff(pre,post))
rawDiffSD <- function(pre,post) sd(rawDiff(pre,post))
percDiffSD <- function(pre,post) sd(percDiff(pre,post))
residDiffSD <- function(pre,post) sd(residDiff(pre,post))
corrDiffTests <- function(pre,post) {
aDF<-data.frame(
rawDiff = rawDiff(pre,post),
percDiff = percDiff(pre,post),
residDiff = residDiff(pre,post)
)
aCor<-cor(aDF)
aList<-as.list(aCor[lower.tri(aCor)])
names(aList)<-combn(colnames(aCor),2,FUN=paste,collapse='_')
aList
}
4. For each iteration, we want to do some things to the DF and record our results.
getPrePostSummary <- function(pre,post,listOfStatsFuncs){
library(plyr)
llply(listOfStatsFuncs,
function(aFunc){
rez<-aFunc(pre,post)
rez
})
}
I want to put the list of functions together so we can pass it to our summary function.
listOfFunc <- list(rawDiffMean, rawDiffSD, percDiffMean, percDiffSD, residDiffMean, residDiffSD, corrDiffTests)
names(listOfFunc) <- c('rawDiffMean', 'rawDiffSD', 'percDiffMean', 'percDiffSD', 'residDiffMean', 'residDiffSD','corrDiffTests')
5. Let’s put it all together.
simRezzies <- ldply(
1:ITERS,
function(x){
aDF <- prePostDFgen(
N,
cortPopMeanT1,
cortPopSDT1,
cortIndivDiffMean,
cortIndivDiffSD,
epsilonM,
epsilonSD,
distInit='gamma',
distDiff='norm')
sumStats <- getPrePostSummary(aDF$pre, aDF$post, listOfFunc)
as.data.frame(sumStats)
})
Here’s a sample of a one of our simulated data sets
testDF<-prePostDFgen(
N,
cortPopMeanT1,
cortPopSDT1,
cortIndivDiffMean,
cortIndivDiffSD,
epsilonM,
epsilonSD,
distInit='gamma',
distDiff='norm')
ggplot(melt(testDF),aes(x=value))+
geom_histogram(aes(fill=variable,y=..density..),position=position_dodge())+
geom_density(alpha=.2, aes(fill=variable))+theme(panel.background=element_rect(fill='#FFFFFF'))
## No id variables; using all as measure variables
## stat_bin: binwidth defaulted to range/30. Use 'binwidth = x' to adjust this.
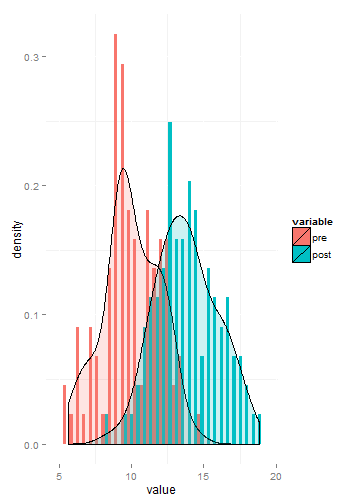
Here’s the result!
simRezzies$RawBias <- expRawDiff-simRezzies$rawDiffMean
simRezzies$PercBias <- expPercDiff-simRezzies$percDiffMean
library(knitr)
kable(simRezzies,digits=2)
| rawDiffMean | rawDiffSD | percDiffMean | percDiffSD | residDiffMean | residDiffSD | corrDiffTests.rawDiff_percDiff | corrDiffTests.rawDiff_residDiff | corrDiffTests.percDiff_residDiff | RawBias | PercBias |
|---|---|---|---|---|---|---|---|---|---|---|
| 4.01 | 1.73 | 0.44 | 0.26 | 0 | 1.62 | 0.87 | 0.94 | 0.67 | -0.01 | -0.04 |
| 4.09 | 1.49 | 0.43 | 0.22 | 0 | 1.42 | 0.87 | 0.95 | 0.69 | -0.09 | -0.03 |
| 3.95 | 1.74 | 0.43 | 0.23 | 0 | 1.55 | 0.92 | 0.89 | 0.68 | 0.05 | -0.03 |
| 4.23 | 1.81 | 0.48 | 0.27 | 0 | 1.70 | 0.85 | 0.94 | 0.65 | -0.23 | -0.08 |
| 4.02 | 1.82 | 0.45 | 0.35 | 0 | 1.67 | 0.80 | 0.92 | 0.59 | -0.02 | -0.05 |
| 4.29 | 1.72 | 0.46 | 0.23 | 0 | 1.64 | 0.88 | 0.95 | 0.71 | -0.29 | -0.06 |
| 4.04 | 1.89 | 0.44 | 0.25 | 0 | 1.81 | 0.88 | 0.95 | 0.71 | -0.04 | -0.04 |
| 4.28 | 1.98 | 0.47 | 0.29 | 0 | 1.86 | 0.87 | 0.94 | 0.68 | -0.28 | -0.07 |
| 3.94 | 1.67 | 0.43 | 0.22 | 0 | 1.66 | 0.86 | 0.99 | 0.79 | 0.06 | -0.03 |
| 3.92 | 1.73 | 0.44 | 0.25 | 0 | 1.69 | 0.89 | 0.98 | 0.78 | 0.08 | -0.04 |
| 4.33 | 1.61 | 0.47 | 0.21 | 0 | 1.61 | 0.79 | 1.00 | 0.79 | -0.33 | -0.07 |
| 4.22 | 1.65 | 0.46 | 0.25 | 0 | 1.58 | 0.85 | 0.96 | 0.69 | -0.22 | -0.06 |
| 3.61 | 1.39 | 0.41 | 0.23 | 0 | 1.33 | 0.80 | 0.96 | 0.62 | 0.39 | -0.01 |
| 3.99 | 1.50 | 0.46 | 0.28 | 0 | 1.39 | 0.84 | 0.93 | 0.63 | 0.01 | -0.06 |
| 3.75 | 1.62 | 0.42 | 0.22 | 0 | 1.51 | 0.89 | 0.94 | 0.71 | 0.25 | -0.02 |
| 3.90 | 1.92 | 0.43 | 0.28 | 0 | 1.83 | 0.83 | 0.95 | 0.66 | 0.10 | -0.03 |
| 4.22 | 1.75 | 0.44 | 0.22 | 0 | 1.74 | 0.87 | 1.00 | 0.82 | -0.22 | -0.04 |
| 4.10 | 1.66 | 0.45 | 0.22 | 0 | 1.64 | 0.90 | 0.99 | 0.82 | -0.10 | -0.05 |
| 4.25 | 1.49 | 0.46 | 0.21 | 0 | 1.45 | 0.87 | 0.98 | 0.75 | -0.25 | -0.06 |
| 3.77 | 1.75 | 0.40 | 0.22 | 0 | 1.74 | 0.87 | 1.00 | 0.84 | 0.23 | 0.00 |
| 4.07 | 1.80 | 0.44 | 0.22 | 0 | 1.76 | 0.89 | 0.98 | 0.80 | -0.07 | -0.04 |
| 3.90 | 1.86 | 0.41 | 0.27 | 0 | 1.80 | 0.88 | 0.97 | 0.76 | 0.10 | -0.01 |
| 4.23 | 1.67 | 0.46 | 0.25 | 0 | 1.60 | 0.88 | 0.96 | 0.72 | -0.23 | -0.06 |
| … |
sumSimRezzies<-as.data.frame(sapply(simRezzies,mean))
kable(sumSimRezzies,col.names='Mean')
| Mean | |
|---|---|
| rawDiffMean | 4.0030541 |
| rawDiffSD | 1.7294373 |
| percDiffMean | 0.4341326 |
| percDiffSD | 0.2447261 |
| residDiffMean | 0.0000000 |
| residDiffSD | 1.6614440 |
| corrDiffTests.rawDiff_percDiff | 0.8648646 |
| corrDiffTests.rawDiff_residDiff | 0.9610976 |
| corrDiffTests.percDiff_residDiff | 0.7275404 |
| RawBias | -0.0030541 |
| PercBias | -0.0341326 |